Pharmacogenomics and Precision Medicine Part 1
Steph’s Note: Is it just me, or has 2023 been the year of new topic areas for tl;dr? We’ve had some phenomenal pediatric content published…we’ve hit critical care hard… And now, because we’re very lucky to have such awesome contributors, it’s time to introduce yet another new topic: pharmacogenomics.
Meet Saba Arshad, PharmD. Saba is currently a practicing pharmacist in Texas, but she actually started her pharmacy life overseas. After completing all the steps to obtain a license in the US, she worked at a specialty pharmacy in Boston, where she learned about URAC. She also worked with hepatitis C, HIV, antipsychotic, and alcohol and opioid abuse patients. She went on to work for local and big box chain pharmacies before deciding to move from retail into consulting. Currently, she serves her community as a pharmacist consultant in the realms of pharmacogenomics and nutrigenomics. Her 'Why' is deeply rooted in mental health.
Take it away, Saba!
Pretty crazy that a mere 1% of our genetic makeup accounts for this and more! (Image)
Have you ever wondered what makes you UNIQUE? As humans, we share ~99.9% of our genetic makeup. But that 0.1% difference defines why some of us have pretty blue eyes or green gray eye color, curly versus straight hair, long versus short eyelashes, and the list goes on…
That 0.1% variability also includes genes that play an important role in determining how our bodies metabolize (process) medications. This is critical for achieving maximum therapeutic outcomes. Precision medicine is an innovative approach that considers individual differences in patients’ genes, environments, and lifestyles. Pharmacogenomics is one part of precision medicine, and one of the major goals is to implement this strategy broadly in medical care – focusing on the right drug at the right dose at the right time for the right patient the very first time.
In simple words, pharmacogenomics (PGx) can inform us how we process over 200+ medications (based on what panels are offered by the labs) with the help of a simple, non-invasive test. (How cool, right?!?!) This is known as pharmacogenomic testing.
Let’s take a quick trip down memory lane…You all remember pharmacokinetics (PK) vs pharmacodynamics (PD), right?
Quick refresher:
PK: What the body does to the medication. The body takes the medication on a journey from being absorbed to the point of being excreted out of the body.
PD: What the medication does to the body. That is, what effect does it produce? It can include anything from relieving pain to the antiplatelet effect of aspirin and more.
People taking the same medication for the same condition can have very different responses:
(Image)
Based on the above information, understanding metabolizer status is of utmost importance. We all have heard our patients saying:
“This medication does not work for me.”
“It gives me rashes, and I just took the first dose! I will never take this one again.”
“I need more of this pain-relieving medication.”
“I want the blue colored pills. Otherwise it does not work for me!”
And the list goes on…
Why this significant variation? The simple reason is that all of us have unique bodies and process medications and environmental toxins at different rates. Generally, there are 4 types of metabolizer statuses:
(Image)
Quick side note: we won’t be going into too much detail about prodrug metabolism. If someone is taking a prodrug, which is an inactive form of drug that requires activation in the liver, it can also be affected by these metabolizer statuses. But as one quick prodrug example…
You have a patient on clopidogrel, who is also a CYP2C19 poor metabolizer. As such, they won’t be as efficient at converting the inactive clopidogrel to its active form compared to a normal metabolizer. This leads to an undesirable lack of effect, which in this case is less antiplatelet effect! Alternative therapy is necessary for optimal antiplatelet protective effects.
Are there evidence-based guidelines for Pharmacogenomics?
Indeed there are! Check out these samples:
CPIC: Clinical Pharmacogenetics Implementation Consortium of the National Institute of Health’s Pharmacogenomics Research Network
The CPIC publishes genotype-based drug guidelines to help clinicians understand how available genetic test results could be used to optimize drug therapy. CPIC is managed at Stanford University and St. Jude’s Research Hospital.
PharmGKB: Pharmacogenomics Knowledge Base
PharmGKB® is a registered trademark of HHS and is financially supported by NIH/NHGRI/NICHD. It is also managed at Stanford University.
The goal of these two organizations is to provide peer-reviewed, updated, evidence-based, freely accessible guidelines for gene/drug pairs. These guidelines facilitate the translation of pharmacogenomic knowledge from research bench to bedside.
DPWG: Royal Dutch Association for the Advancement of Pharmacy - Pharmacogenetics Working Group
Warning… this site is actually in Dutch. So unless you’re up on your Dutch or have time to Google translate, it could be a little more difficult to use. But they are an important and well-recognized source of PGx information across the world.
Other international professional societies include the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) and the French National Network of Pharmacogenetics (RNPGx).
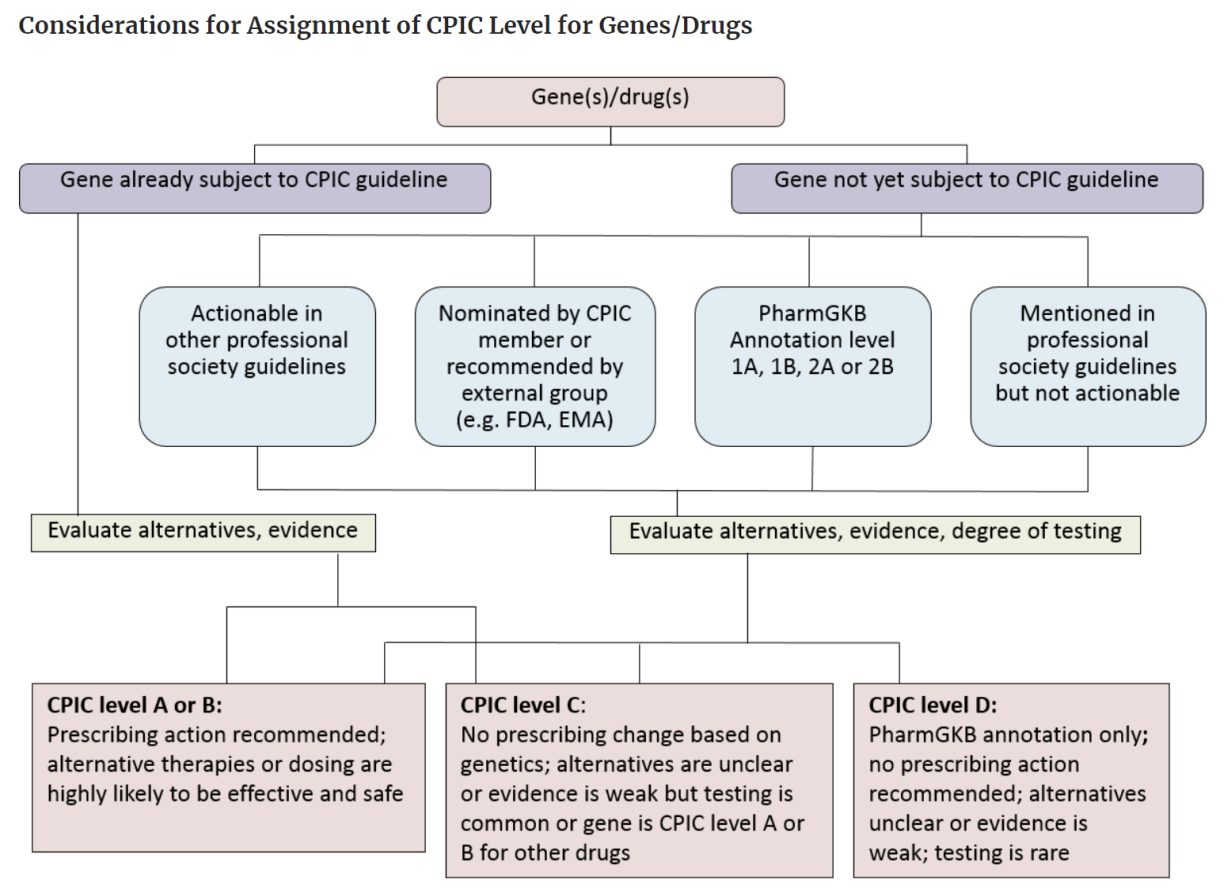
Levels of Evidence in Pharmacogenomics Guidelines
The level of evidence determines if the prescribing actions are recommended.
In CPIC, Level A or B means that they have sufficient evidence for at least one prescribing action so it could be recommended. Look at the chart below.
(Image)
PharmGKB levels of evidence are based on multiple criteria including replication, statistical significance, and study size. Level 1A and Level 1B are considered to be more authentic (in simple words) so they can used for any prescribing action.
The FDA has also published a list of Pharmacogenomic Biomarkers in Drug Labeling, which contains pharmacogenomic information on over 200+ medications to assist clinicians in prescribing. The NIH also has a number of PGx initiatives, including building the Clinical Genome Resource, which is a central clinical resource to better translate precision medicine into practice.
Another online resource is the PharmGKB Pediatric Dashboard. There are currently 399 drugs on the dashboard! This resource also denotes which medications are listed in the Best Pharmaceuticals for Children Act (BPCA).
Applications of Pharmacogenomics
As you can see from the graph below, this section could legitimately be the most ginormous post on this site to date. But instead, it is tl;dr, so I’m going to try to give you an intro to a smattering of PGx therapeutic areas.
This graph illustrates the number of clinically relevant drug-biomarker pair counts per therapeutic area. Of note, the 18 biomarkers included in this chart (in the legend on the right) are only those that impact at least 5 drugs each, so it’s not even an all inclusive list! Also, check out the insane biomarker diversity of the oncology therapeutic class. (Image)
Pharmacogenomics in Psychiatry
Antidepressants are one of the most prescribed drug classes in the United States. In an estimated 30% to 50% of patients, initial antidepressant drug therapy fails because of ineffectiveness or drug-induced adverse effects. Important CYP enzymes are as follows:
CYP2D6: Located on the 22nd chromosome, it has a high degree of allelic variability (over 130 have been identified). It’s involved in the primary metabolism of more than 70 medications with antipsychotics and ADHD medications being the main ones.
Calculating activity score and phenoconversion is a topic for next time.
CYP2C9: This is a phase I drug-metabolizing cytochrome P450 (CYP450) enzyme isoform that plays a major role in the oxidation of both xenobiotic and endogenous compounds. For example: phenytoin.
2C9 plays a major role in metabolism of anticonvulsants and other medications’ pathways as shown below:
CYP2C19: Involved in the metabolism of ~10% of medications. The poor metabolizer (PM) phenotype is important in patients taking TCAs, citalopram, escitalopram, and sertraline.
HLA-A and HLA-B: HLA genes are highly polymorphic, and different alleles play an important role in our immune system. A prominent example is the widely used antiepileptic drug, carbamazepine. The HLA-B*1502 allele is strongly associated with the risk of developing Stevens–Johnson Syndrome (SJS) with carbamazepine, and it is recommended to screen patients with Asian ancestry for the presence of HLA-B*1502 before initiating therapy.
Pyschiatric medications with pharmacogenomic prescribing guidelines as of April 2021. (Image)
Pharmacogenomics in Cardiology
Clopidogrel: This antiplatelet drug belongs to the group of P2Y12 inhibitors. It is commonly prescribed to reduce the risk of myocardial infarction (MI) and stroke in patients with acute coronary syndromes (ACS) and/or following percutaneous coronary intervention (PCI), although newer agents are now available in the market.
Clopidogrel is a thienopyridine prodrug that requires hepatic biotransformation to form an active metabolite. This active metabolite selectively and irreversibly inhibits the purinergic P2Y12 receptor, thereby inhibiting platelet aggregation for the platelet’s lifespan (~10 days). Only 15% of the prodrug is available for transformation to the active agent. The remaining 85% is hydrolyzed by carboxylesterase-1 (CES1) to inactive forms.
Conversion of clopidogrel to its active metabolite requires two sequential oxidative steps involving several CYP450 enzymes (e.g., CYP1A2, CYP2B6, CYP2C9, CYP2C19, and CYP3A4/5). However, in both steps, CYP2C19 has the greatest contribution of all of these enzymes. As such, variability in CYP2C19 metabolism can have HUGE impacts on the effectiveness of clopidogrel’s antiplatelet protection.
We all know about the famous lawsuit Sanofi and Bristol Myers Squibb went through, from which they were ordered to pay $834 million to the state of Hawaii over the Plavix warning label. So kind of a big deal. In my case, I am a rapid metabolizer of CYP2C19, and I know clopidogrel would work for me at a standard dose if I ever suffer a stroke (God forbid!).
Warfarin: This is a widely used anticoagulant with a narrow therapeutic index and large interpatient variability in the dose is required to achieve target anticoagulation. Common genetic variants in CYP2C9, VKORC1, CYP4F2, and the CYP2C cluster (e.g., rs12777823), plus known nongenetic factors, account for 50% of warfarin dose variability. (So how do we get it right?? Check out this all things warfarin post!)
Statins: In the case of statins, the PGx association with the most robust evidence is SLCO1B1. SLCO1B1 encodes for a solute carrier transport molecule involved in the liver intake of statins, and variants of this gene have been associated with statin-associated muscle symptoms (SAMS). One specific variant, rs4149056, can lead to increased exposure to statins outside the liver and has been repeatedly associated with SAMS.
Pharmacogenomics in Oncology
Seriously, there’s not enough time in the world for #allthethings here. But a brief list would include 5-FU, capecitabine (Xeloda), thioguanine, (irinotecan) Camptosar, cisplatin, and more.
One notable example is an interaction between the DPYD gene for the dihydropyrimidine dehydrogenase (DPD) enzyme and fluoropyrimidines (i.e., capecitabine and fluorouracil). Patients with low DPD enzyme activity cannot effectively metabolize these drugs into inactive forms, which can result in life-threatening toxicities. Unfortunately, there have been several fatalities due to this enzymatic deficiency, and there is currently debate about appropriate initiation strategies for these medications.
In addition to these major therapeutic areas, PGx also plays heavily into…
Pain management
Infectious diseases
Rheumatology, and more!
(Btw, yes, I know this is quite the teaser… So if you want to know more about these and other therapeutic areas for which PGx has found utility, you’ll just have to stay tuned for my Part 2 post! Coming soon!!)
What is the process to have a pharmacogenomics test completed?
For such a complex and detailed topic, the process of completing a PGx test itself is actually relatively simple. The testing kit is somewhat like what we have used for COVID-19 testing: a Q-tip shaped cheek swab stick, with colored solution in a test tube for keeping the sample stable during its journey from the patient’s home to the lab’s testing area. Most kits (at least what I have experienced) always have a prepaid shipping label.
There are 2 ways to take your sample:
Cheek swab: A patient can gently swab the gum line or inside cheek area 30 times on one side and then repeat it on the other side. Dip the swab stick into the test tube solution and close the test tube. Put everything back in the kit, and their sample is ready to be analyzed by the lab.
Saliva sample: The container where a person has to spit comes in the test kit from the lab. Fill it up to the mark with saliva, and seal it in the given ziplock-type bag. Put everything back in the kit, and ship the completed kit via the carrier.
What’s the turnaround time for receiving results?
Waiting to know what medication would be best for you and how you will metabolize future medications is exciting but a little scary too! (It sure was for me!) It usually takes 7-10 business days, perhaps up to 15 days, for the results to come in. This really depends on the lab and their testing capacity. But it’s pretty cool to know that in just a week or 2, you can have all this information at your fingertips.
Is Pharmacogenomics the silver bullet to patient care?
Unfortunately… Nope. It’s still just one piece of a puzzle. There is no overnight magical potion that works, making everything better in the blink of an eye. We must consider the epigenetic factors as well as so many other factors, such as liver disease, kidney function, age, sex, etc.
Right Drug Dose Now Act
The “Right Drug Dose Now” Act was introduced by Representatives Tom Emmer (MN-06) and Eric Swalwell (CA-15) in February 2022. The purpose is to better enable implementation of pharmacogenetic testing to prevent adverse drug events and encourage the use of therapies tailored to a patient’s genetic makeup.
The NIH also has other PGx initiatives, which include building the Cinical Genome Research. The key goal of the organization is to build a genomic platform, so patient care improves.
Why does the government care?
Of course, we hope they care because, ahem, quality of human life etc, but also… Do you know how much we WASTE?!!?
Non-optimized medication management (e.g., adverse reactions, inadequate or unnecessary therapies, nonadherence and incorrect dosing) leads to poor patient outcomes. It also takes more time for the clinician and quite frankly, costs a boatload of money. In total, our current (largely non-PGx) trial-and-error approach to medication therapy results in the loss of 275,000 lives and $528.4 billion in total US health care expenditures - each year.
Knowing this, why hasn’t pharmacogenomics taken off like T-Swift yet?
Loooots of reasons. There are still a number of looming barriers to PGx implementation at this point:
Lack of provider knowledge in general but also specifically for interpretation of PGx results
Patient fears of cost and/or being subject to discrimination based on the results
Most people of non-European ancestry are not included in research studies, so lower applicability to these groups
Limited (if any) insurance coverage
Despite these barriers, the future of PGx is not coming or evolving. It is already here! It just keeps on getting better. Wouldn’t you like to know before suffering a stroke that clopidogrel is the antiplatelet for you? Or whether an alternative is required? It saves money, time, and heartache in the long run, not just for our patients but also for healthcare providers and the health system.
If you’re sufficiently hooked on PGx now, be on the lookout for Part 2 of this series, in which I’ll give more details about applications for specific drugs and genes!